Recently, Dhuli's paper and supplementary data have become controversial on Twitter (X). The theme of the Dhuli's paper is "Presence of viral spike protein and vaccinal spike protein in the blood serum of patients with long-COVID syndrome."
On the other hand, the Dhuli's supplementary data teaches that the vaccine-derived DNA was detected in the blood sample through the Nested PCR involving three PCRs, and has led to a huge technical controversy.
In regard to the supplementary data, there is some consideration (speculation) that it is not the Nested PCR since the same primers were used in all three PCRs.
However, is this consideration really correct?
In this connection, Dhuli teaches on page 4 of the paper that the supplementary data follows the protocol disclosed in Markus Aldén's paper published in 2022.
The Aldén's paper is (ref. 1) in the Dhuli's supplementary data, and corresponds to (ref. 18) in the Dhuli's paper. Many seem to have overlooked the Aldén's paper, but isn't it hard to accurately comprehend the Dhuli's documents without referring to this paper?
The Aldén's paper publicizes the possibility of reverse transcription of DNA from the vaccine-derived spike mRNA to the liver cell genome using an "in-vitro test," and this caused a huge ripple due to its experimental conditions.
The supplementary data this time discloses an extension research with regard to the Aldén's paper, and is understood to be an attempt to confirm the same phenomenon through an "in-vivo test" based on the idea of that paper.
In the following discussion, Aldén's paper shall be referred to as "protocol (ref. 1)".
The supplementary data includes 1st PCR, 2nd PCR, and 3rd PCR as the Nested PCR. Interestingly, Dhuli used the different primers from the primers of the protocol (ref. 1), whereas the testing conditions were almost identical to those of the protocol (ref. 1).
Therefore, we need to carefully interpret the Dhuli's Nested PCR and their intentions based on the disclosures of the protocol (ref. 1), and precisely question to the description of the Dhuli's Nested PCR.
The forward and reverse primers used in the PCR in the protocol (ref. 1) are shown in its Table 1, and they can be shown as the attached FIG. 1 by using the amplicon shown in its Table 2. The amplicon size of the protocol (ref. 1) is 444bp. In the protocol (ref. 1), the DNA amplicon verified by the Sanger sequencing is 100% identical to the vaccine sequence.
Forward primer
CGAGGTGGCCAAGAATCTGA
Reverse primer
TAGGCTAAGCGTTTTGAGCTG
On the other hand, the forward and reverse primers used in the Dhuli's Nested PCR are shown in the supplementary data, and they can be shown as the attached FIG. 2 by using the amplicon shown in Table 2 of the protocol (ref. 1).
Forward primer
CGAGGTGGCCAAGAATCTGA
Reverse primer
TCTGGAACTAGCAGAGGTGG
That is, the forward primer in the supplementary data is consistent with that of the protocol (ref. 1), while the reverse primer in the supplementary data is positioned inside that of the protocol (ref. 1).
The range (size) of the primers in the supplementary data is 400 bp, which is 44 bp shorter than the range (= 444 bp) of the primers in the protocol (ref. 1).
Did Dhuli perform the Nested PCR including three PCRs using shorter primers different from those of the protocol (ref. 1)? Can't be considered that the primers of the protocol (ref. 1) were used in the 1st PCR and the shorter primers were used in the 2nd and 3rd PCRs in the Dhuli's Nested PCR?
The Nested PCR in which only one primer (in this case the reverse primer) is positioned inside the other is known as a Semi Nested PCR.
In the PCR Analysis section of the supplementary data, quotation mark (1) indicates that the PCR was performed using the specific primers disclosed in the protocol (ref. 1) (see red marker). This section also teaches the forward and reverse primers that are shorter than those of the protocol (ref. 1) (see blue marker). Furthermore, this section teaches the experimental conditions to be applied to all PCRs (see yellow marker).
On the other hand, the Nested PCR section of the supplementary data teaches that the same primers and conditions were used for both 2nd and 3rd PCRs (see green marker). However, this section does not teach that the 2nd and 3rd PCRs were performed using the same primers as that of the 1st PCR.
Therefore, these statements suggest that the primers of the protocol (ref. 1) were used for the 1st PCR, and shorter primers were used for the 2nd and 3rd PCRs.
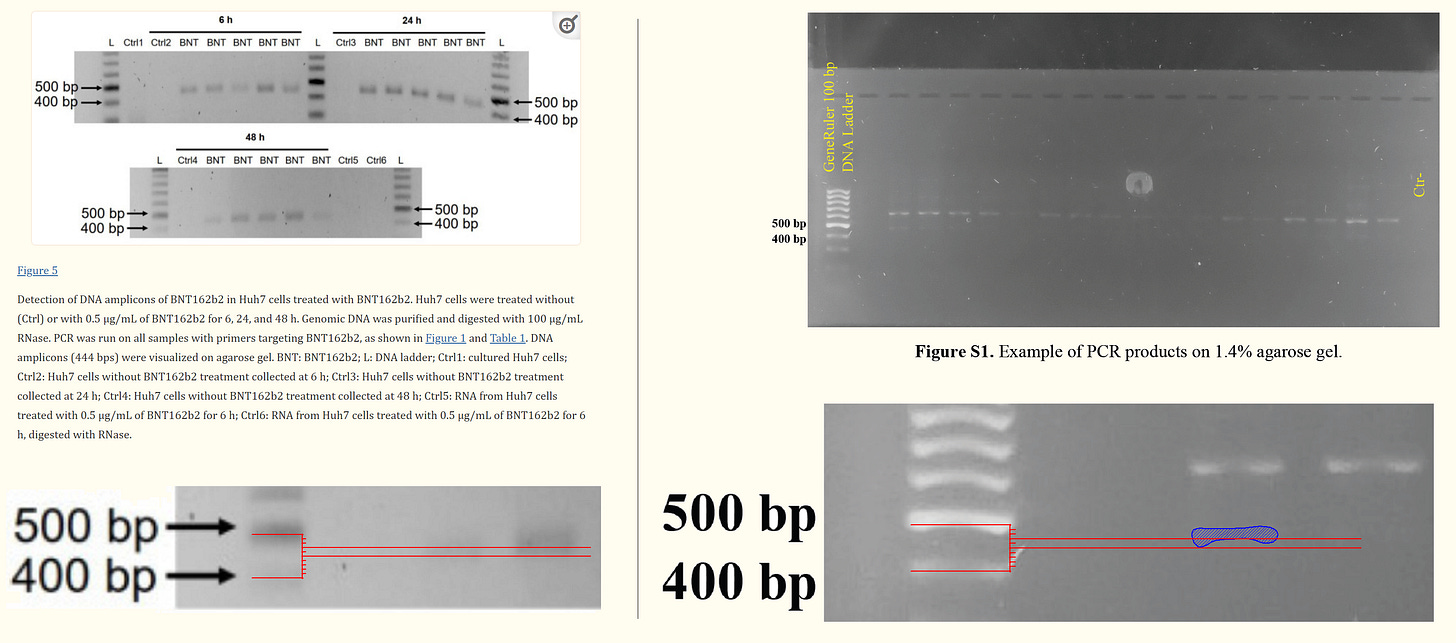
In the section of PCR Analysis in Supplementary Results, it is disclosed that "A band of approximately 440 base pairs (bp) was detected, which corresponded to the expected size of the fragment amplified by the BNT162b2 primers (see purple marker)."
However, referring to Figure. S1 of the supplemental data, the obtained band size was not 440 bp, but approximately 470 bp. On the other hand, the result of the protocol (ref. 1) is also approximately 470bp, which is consistent with the result of the Figure. S1 in the supplementary data.
Therefore, in the Dhuli's supplementary data, there is no contradiction in interpreting that the primers of the protocol (ref. 1) were used in the 1st PCR, and short primers were used in the 2nd and 3rd PCRs. Not only that, the experimental results of the supplementary data can be interpreted smoothly.
This post is not intended to blame anyone and does not question the validity of the Dhuli's paper and supplementary data. In fact, the possibility that the three PCRs were performed using the same primers cannot be ruled out.
Therefore, we need to know the detailed protocol of the Dhuli's Nested PCR. If anyone tries to contact to Dhuli, I would like you to ask the above questions . (anyone may have already done though…)
Dhuli's email address: Corresponding Author: Kristjana Dhuli, MSc;
e-mail: kristjana.dhuli@assomagi.org
Thank you.
Reference lists
Dhuli's paper
https://www.europeanreview.org/wp/wp-content/uploads/013-019-2.pdf
Dhuli's supplementary data
https://www.europeanreview.org/wp/wp-content/uploads/supplementary-data.pdf
Aldén's paper(protocol (ref. 1))